Overview
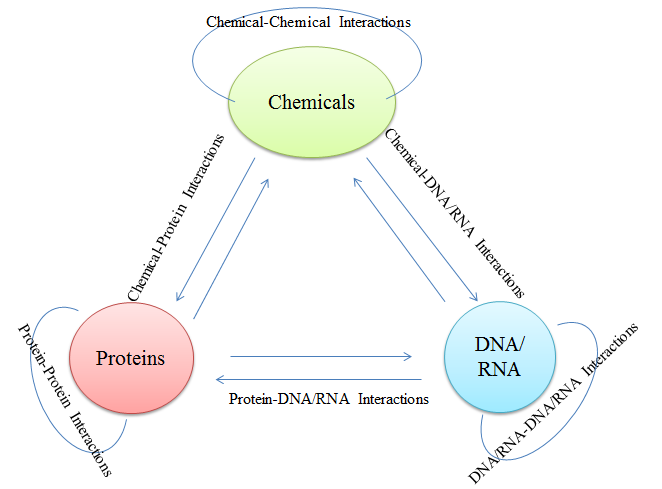
BioTriangle is a feature-rich toolkit used for the characterization of various complex biological molecules and interaction samples, such as chemicals, proteins, DNA/RNA, and their interactions. Biotriangle calculates nine types of features including chemical descriptors or molecular fingerprints, structural and physicochemical features of proteins and peptides from amino acid sequence, composition and physicochemical features of DNA/RNA from their primary sequences, chemical-chemical interaction features, chemical-protein interaction features, chemical-DNA/RNA interaction features, protein-protein interaction features, protein-DNA/RNA interaction features, and DNA/RNA-DNA/RNA interaction features. These chemical descriptors from chemicals have been routinely used in QSAR/SAR, virtual screening, database search, ranking, drug ADME/T prediction and other drug discovery processes. These sequence-derived structural and physicochemical features have been widely used in the development of machine learning models for predicting protein structural and functional classes, post-translational modification, subcellular locations and peptides of specific properties. These composition and physicochemical features of DNA/RNA have been routinely used in several problems in computational genomics and genome sequence analysis. Those interaction features from crossover of different molecular types enable the users to conveniently study various complex molecular recognitions. We recommend BioTriangle to analyze and represent various complex molecular data under investigation. We hope that the webserver will be helpful when exploring questions concerning structures, functions and interactions of various molecular data in the context of systems biology.
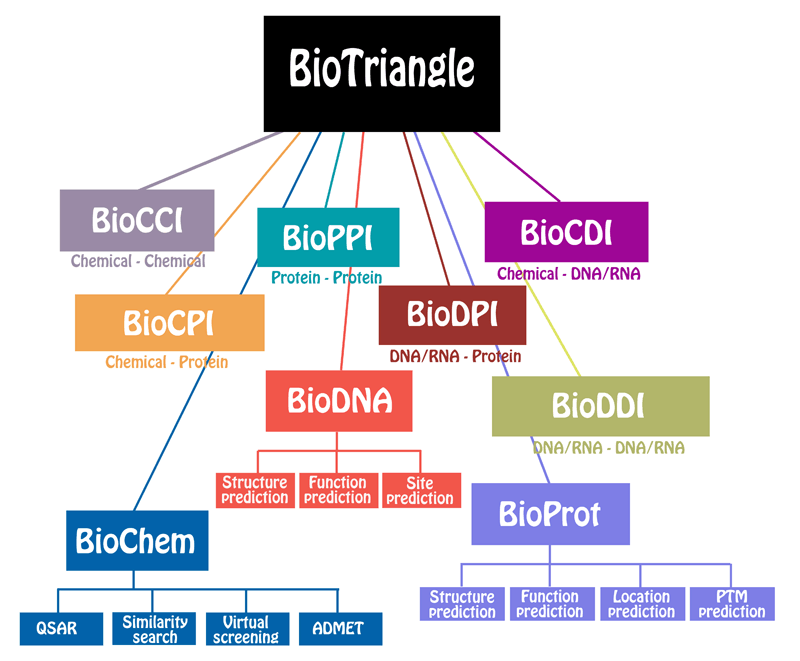
| Tools | Features | Description |
|---|---|---|
| BioChem |
|
Details>> |
| BioProt |
|
Details>> |
| BioDNA |
|
Details>> |
|
||
|
||
| BioCCI BioDDI BioPPI |
|
Details>> |
| BioCDI BioCPI BioDPI |
|
Details>> |
BioChem, BioProt, BioDNA
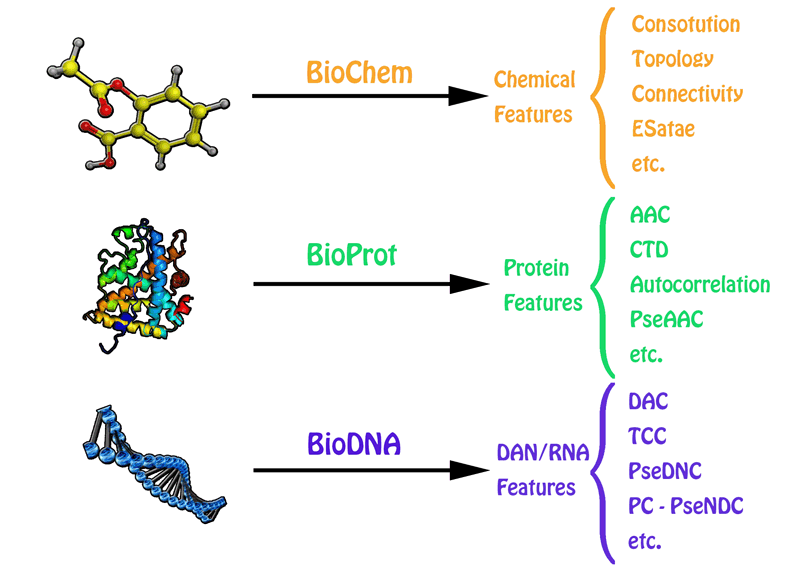
- A: The chemical features of small molecules could be calculated through BioChem tool from BioTriangle.
- B: The sequence-derived structural and physicochemical features of peptides and proteins could be calculated through BioProt tool from BioTriangle.
- C: The composition and physicochemical features of DNA/RNA could be calculated through BioDNA tool from BioTriangle.
BioCCI, BioPPI, BioDDI
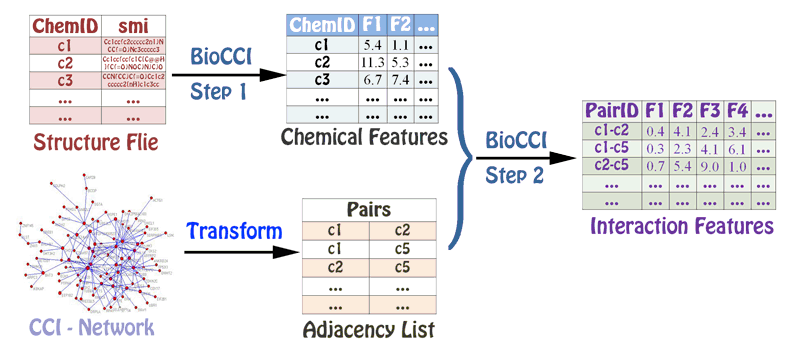
The calculation process for BioCCI, BioPPI and BioDDI is similar to each other.
- Step 1: The molecular structures or sequences of the associated chemicals, proteins, and DNAs/RNAs in the chemical-chemical, protein-protein, and DNA/RNA-DNA/RNA interaction networks are provided to calculate the corresponding molecular features.
- Step 2: The adjacency list file and the molecular features in Step 1 are provided to calculate the final interaction features.
BioCPI, BioDPI, BioDCI
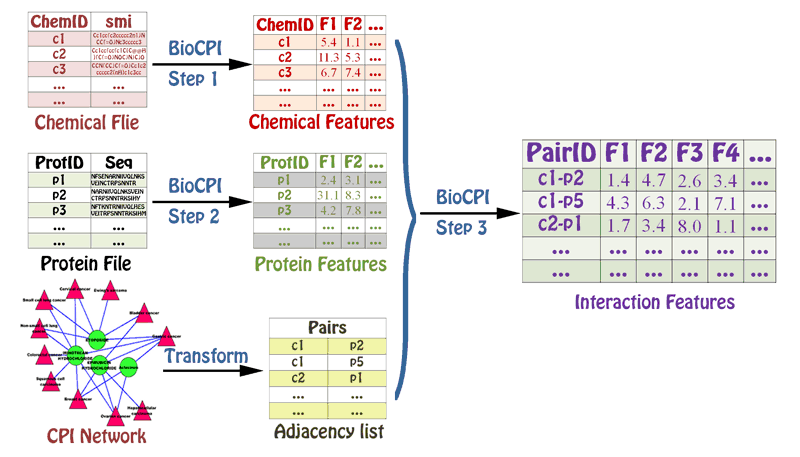
The calculation process for BioCDI, BioCPI and BioDPI is similar to each other.
- Step 1: The molecular structures or sequences of the associated chemicals, proteins, and DNAs/RNAs in the chemical-chemical, protein-protein, and DNA/RNA-DNA/RNA interaction networks are provided to calculate the corresponding molecular features.
- Step 2: The molecular structures or sequences of another molecular object from interaction data are provided to calculate the corresponding molecular features.
- Step 3: The adjacency list file and the molecular features in Step 1 and 2 are provided to calculate the final interaction features.